What are residue communities
Evolutionary information is conserved in naturally occurring proteins, and it plays a significant role in specifying their three-dimensional structure and function. An approach is developed to identify the evolutionary signatures (highly ordered networks of coupled amino acids, termed "residue communities", RCs) from protein sequence profiles.
How to compute residue communities
Pairwise couplings
A pairwise coupling is a pattern between two covariant amino acids, and those couplings are conserved across evolution due to the functional and structural constraints. They can be used to predict protein structures, binding interfaces and protein-protein interactions, as well as protein design.
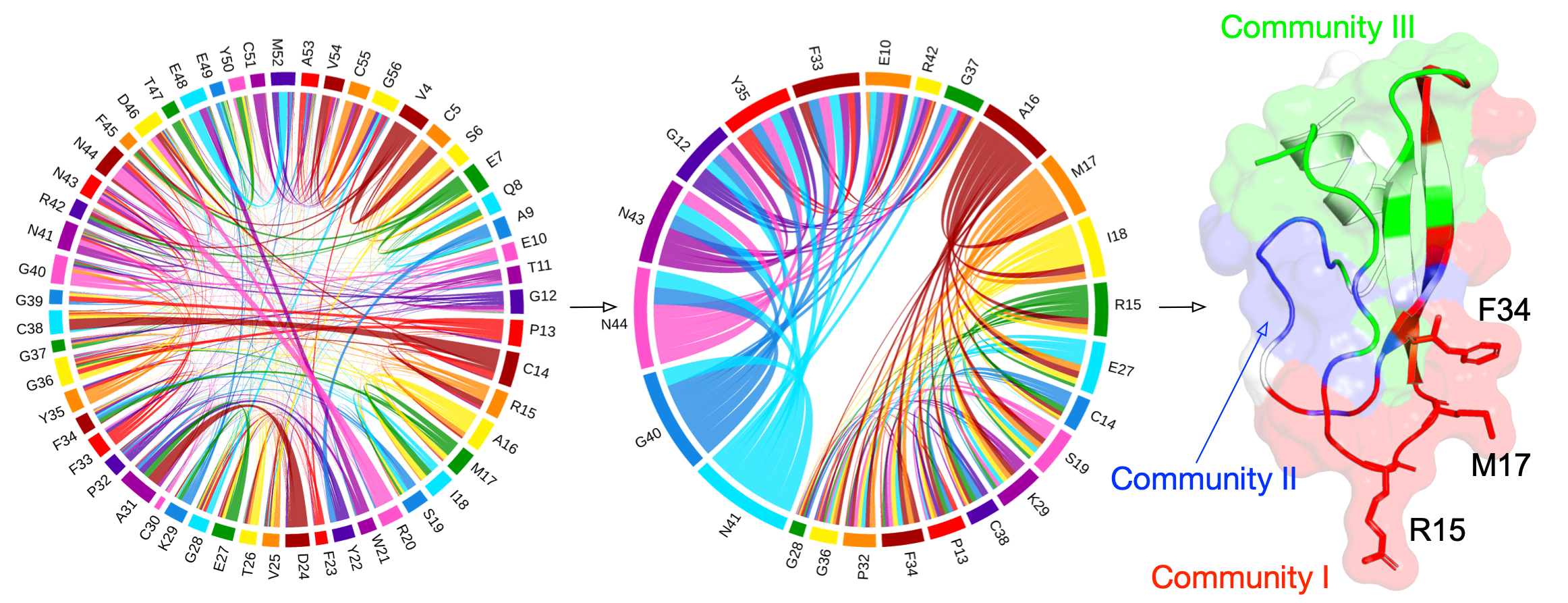
Residue communities
The couplings between pairwise amino acids are analyzed using the principles in the SAEC method1 to make the residue communities as much statistically independent as possible. The principles are as follows:
- community I consists of residues at the \(i\)th position of \(\mathbf{v}_{k=2}^i>\text{max}(\mathbf{v}_{k=4}^i, \epsilon)\),
- community II includes residues at the \(i\)th position of \(\mathbf{v}_{k=2}^i<-\text{max}(\mathbf{v}_{k=4}^i, \epsilon)\),
- community III includes residues at the \(i\)th position of \(\mathbf{v}_{k=4}^i>\text{max}(\mathbf{v}_{k=2}^i, \epsilon)\).
We use an \(\epsilon=0.05\) as a threshold to project the amino acids reduced from the coupling matrix and extract meaningful residue communities.
Mapping to structure
The computed RCs are mapped to the tertiary structure (PDB ID: 1AAP).

Function-relevant sites
A conformational change of the G-loop make an extended structure to plug into the head region of the scMdm12Δ and cover the solvent-exposed concave surface of scMdm12Δ2. The SAEC method reflects that residue community (in red) is relevant to the conformational change, the two structures of Mmm1 as single domain and in the complex are aligned to each other for comparison. The statistical qualities of the residue communities (in green) do not capture the important information from the G-loop when Mmm1 adopts a shape as a monomer, while the Mmm1 changes its shape via the G-loop (residue community in red) in response to the formation of a complex between the Mmm1 and Mdm12 proteins. Read More >>

References
- [1] N. J. Cheung et al., Comput. Struct. Biotechnol. J. 19, 3556–3563 (2021)
- [2] T. R. Hynes, et al., Biochemistry 29, 10018-10022 (1990)
- [3] H. Jeong, J. Park, Y. Jun, C. Lee, PNAS 114, E9502–E9511 (2017)