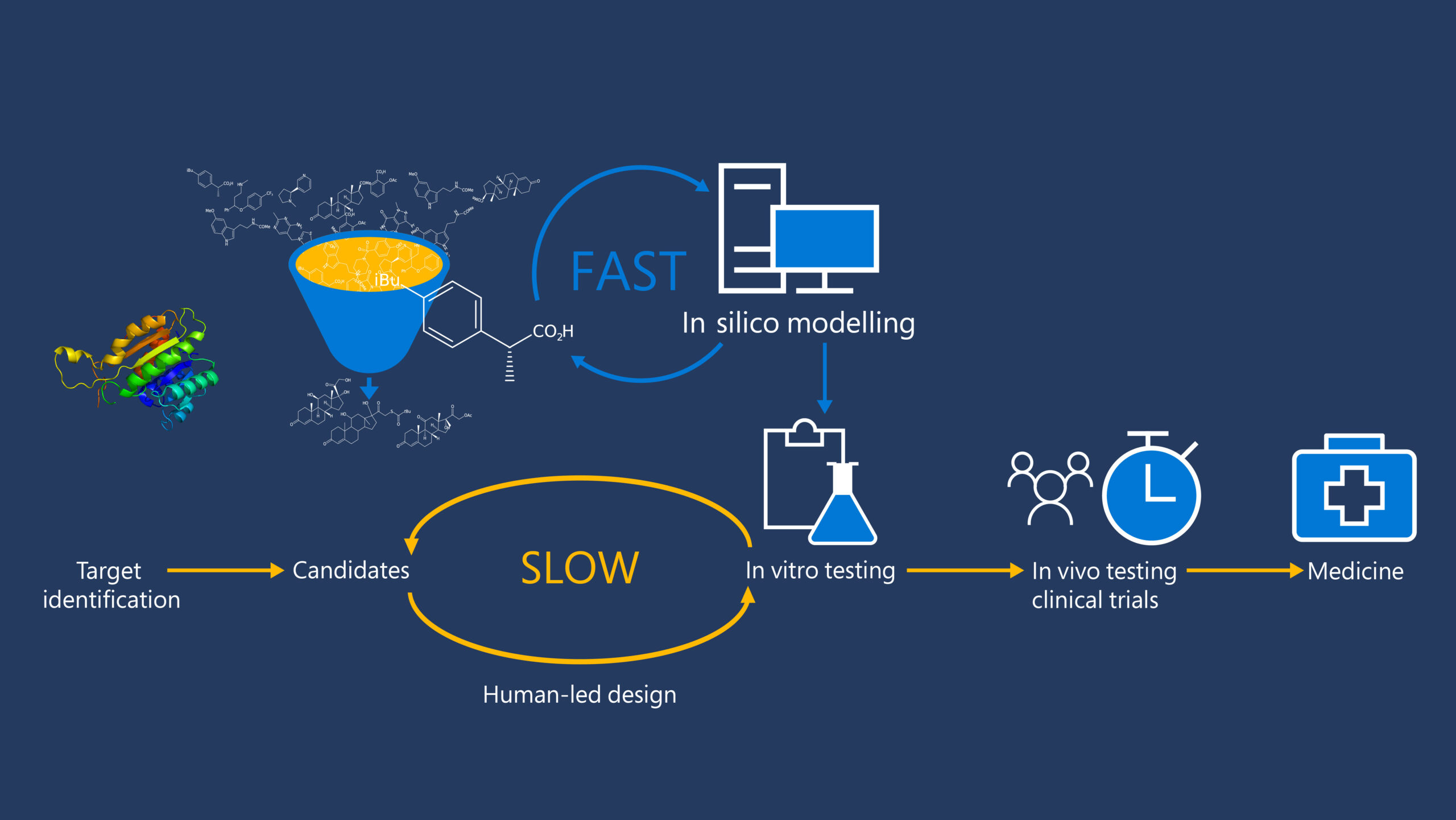
Text Text Text Text Text Text
Text Text Text Text Text Text.
Text Text Text Text Text Text.
- Text Text Text Text Text Text
- Text Text Text Text Text Text
- Text Text Text Text Text Text
Text Text Text Text Text Text
Leveraging pyhysics-informed algorithms, we seek to understand the fundamental principles evolved in protein structure and function, to encode these principles into computer programs, and to use them to address some of the most complex and interesting challenges in biology.
We are always looking for talented, enthusiastic, and diverse scientists to join our efforts. Our philosophy is that the more closely interacting a research group is, the better the science and the more fun overall. We highly encourage you to join us if you have any relevant backgounds as follows,
Postdoc positions are potentially available, including Fellowships in Computational Biology and Bioinformatics. Please contact us if interested.
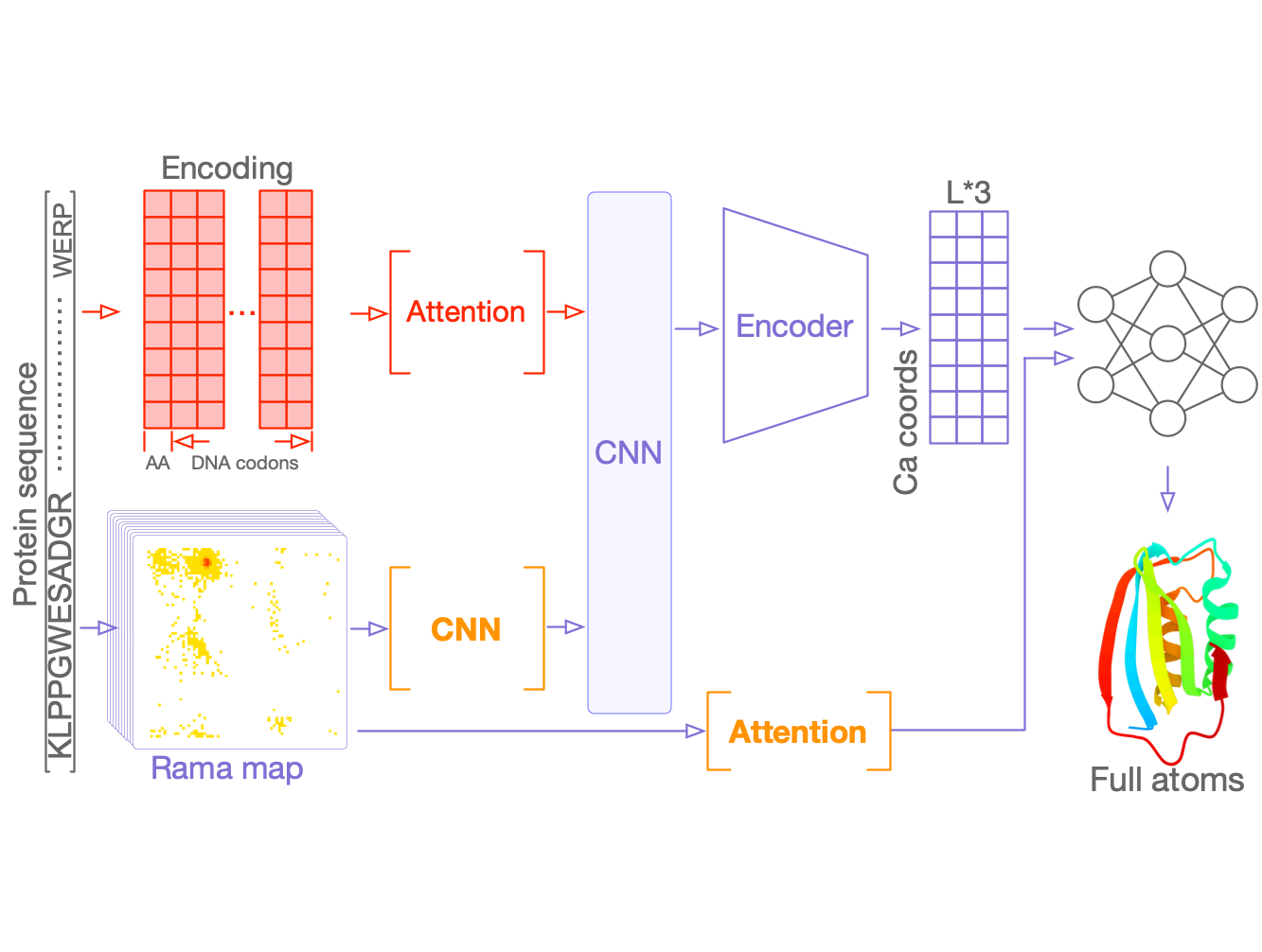
Learn to design protein: Protein sequences contain essential evolutionary information that specifies their structures and functions. We are developing methodologies that utilize the signatures derived from this information and aim to leverage them in designing proteins with specific functions.
Protein-based therapeutics: Machine learning has emerged as a powerful tool for advancing the development of protein therapeutics in medicine, leading to a revolution in the field of drug discovery. Specifically, we are developing novel algorithms to design proteins with (1) enzymatic or regulatory activity, (2) specific targeting activity, and (3) vaccines. The ultimate goal is to engineer these proteins to treat various of human diseases, thereby offering new treatmentis for patients in need.
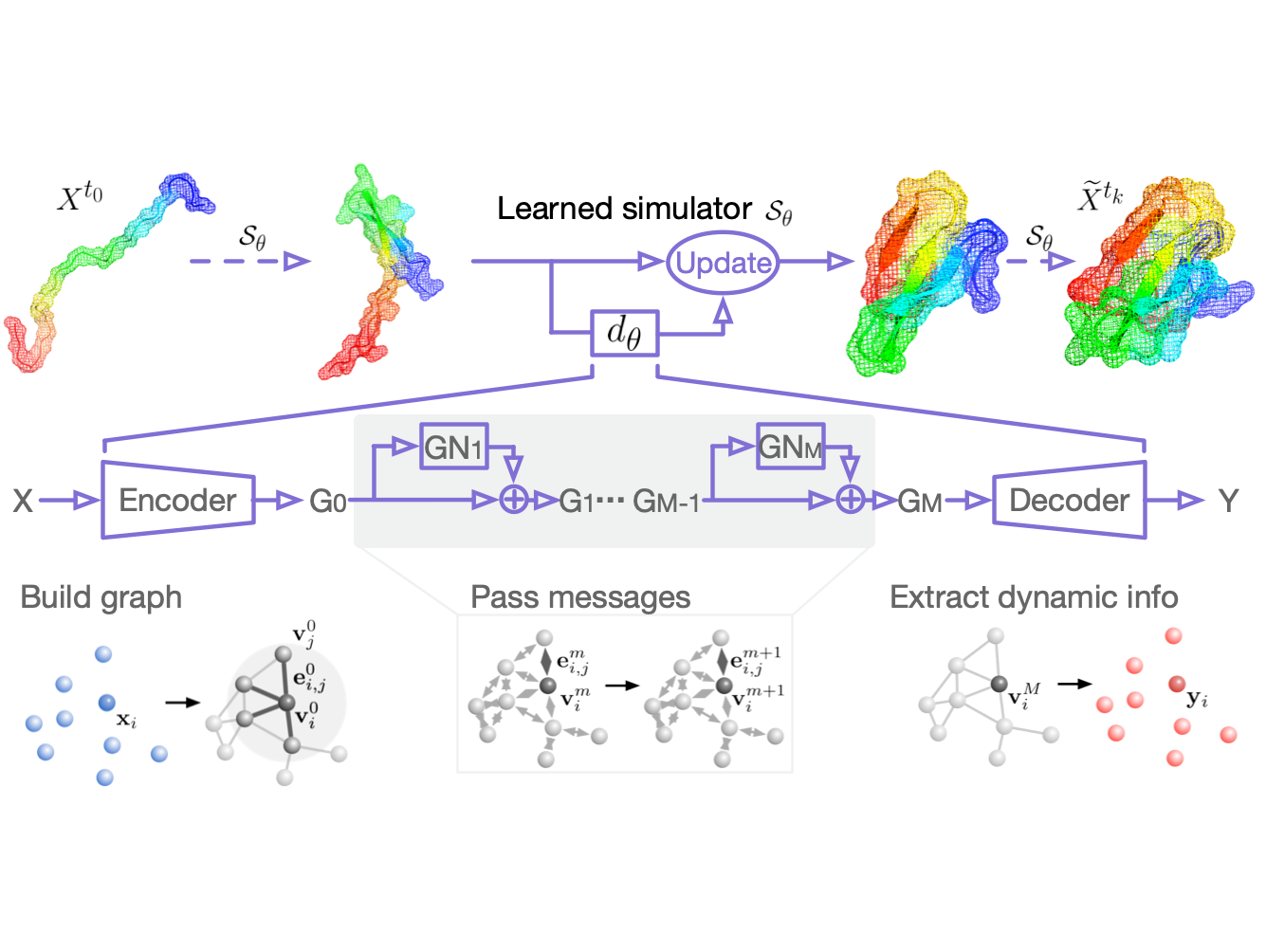
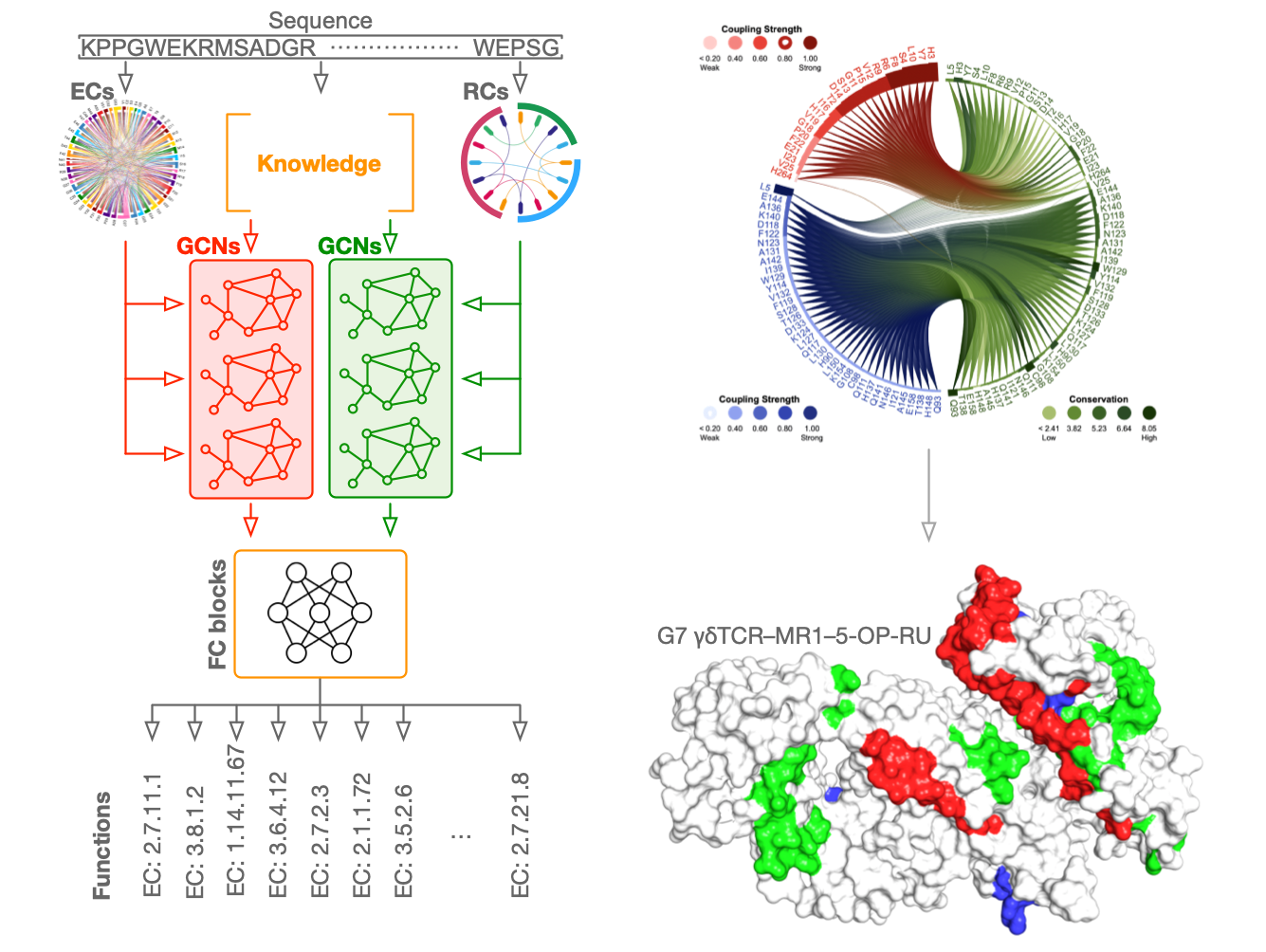
Protein function annotation: we develop physic-informed learning-based methods to characterize protein functional sites at the residue level. Protein function is exquisitely dependent on compactly folded structures that combine energetic stability with intrinsic flexibility. We are now trying to define the thermodynamic and evolutionary origins of metamorphic folding.
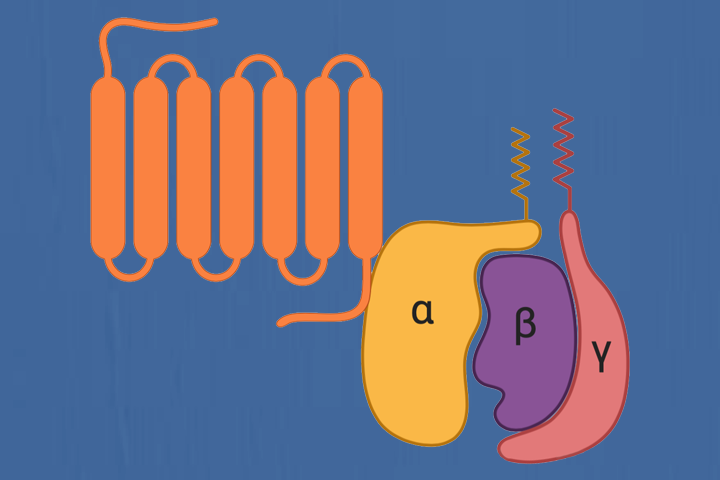
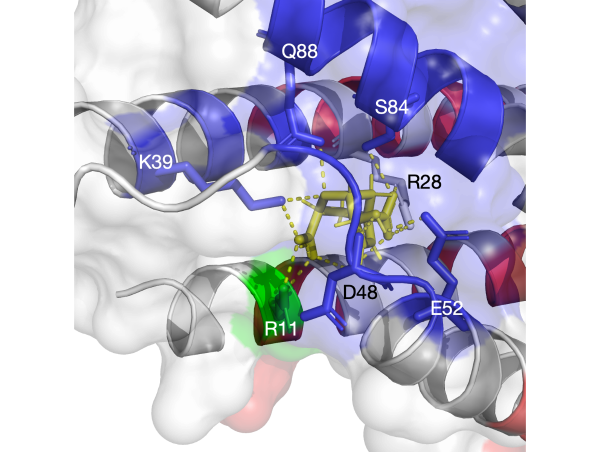
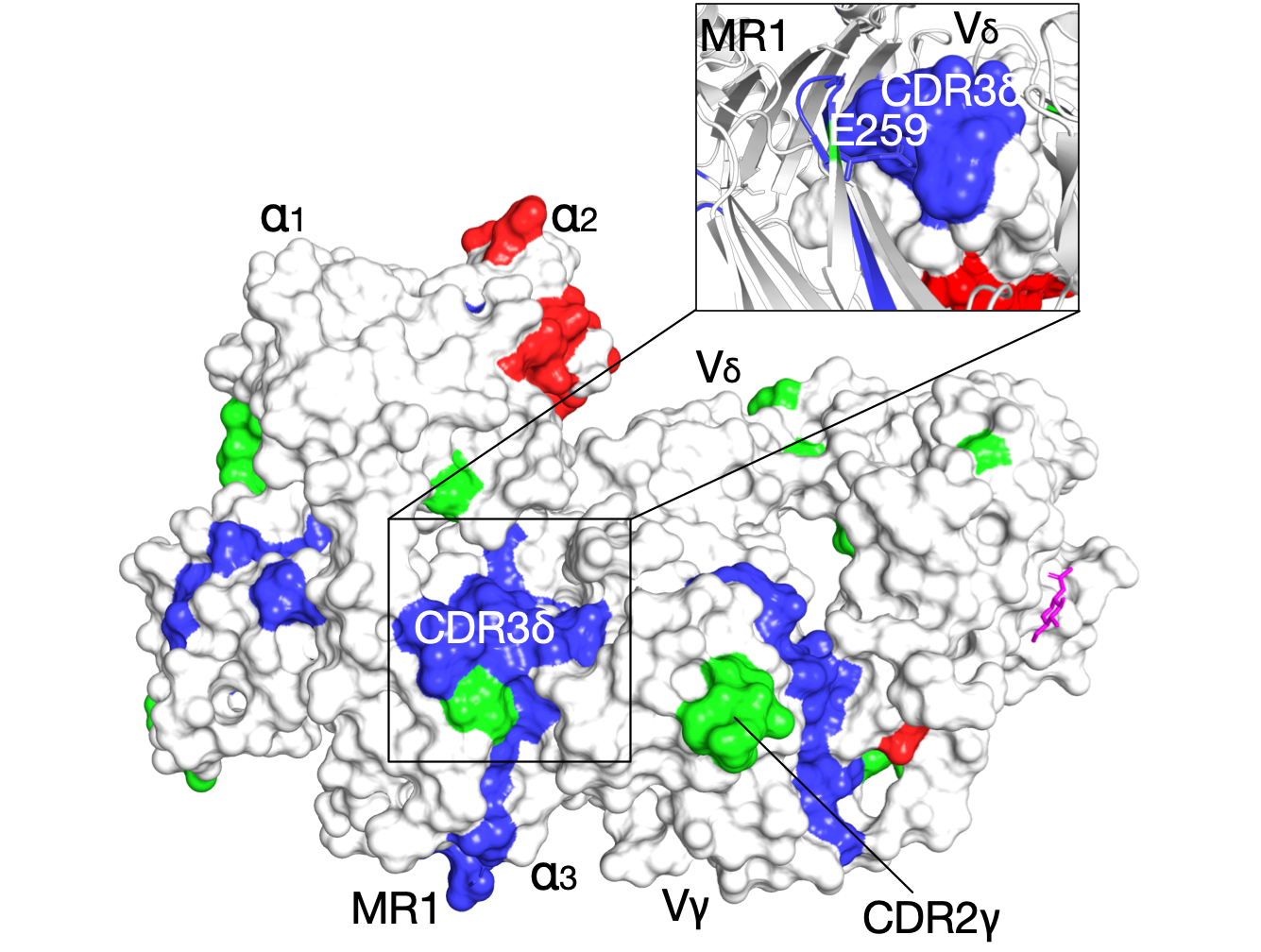
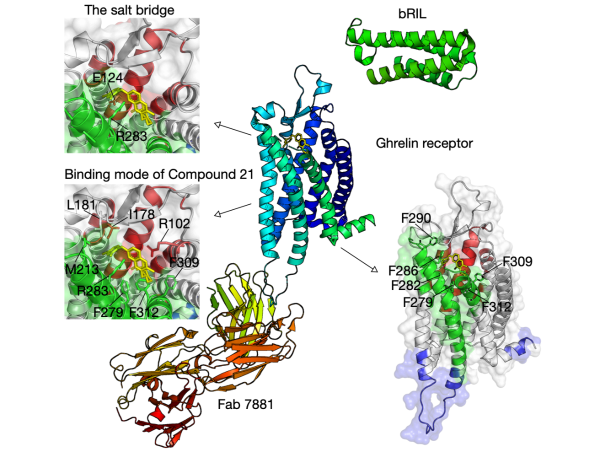
Molecular recognition: the interation (promoting or disrupting) between two proteins often controls biological signals. We leverage computational approaches on three systesms: (1) binding mechanisms of GPCR-G protein; (2) T cell receptor (TCR) activation, important in the defense against cancer; and (3) chemokines, directly implicated in human diseases.

Physics-informed learning: we develop explainable artificial intelligence using information obtained by enforcing the physical laws.
Probabilistic deep learning: we build deep learning-based models with probabilistic modules for representing and processing uncertainty in both data and models.
Quantum mechanics in biology: we seek to model biological interactions in light of quantum mechanical effects and understand aspects of biology that cannot be accurately described by the classical laws of physics.
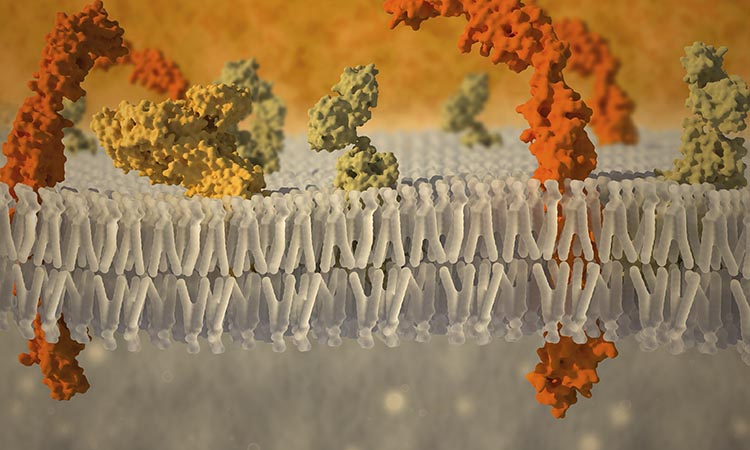
Lipid transport proteins: intestinal lipid transport is a multistep process that requires the coordinated regulation of a series of pathways that lead to the transport of lipolytic products and micellarized lipid across the brush-border membrane, through vectorial delivery through the apical cytoplasmic compartment to the endoplasmic reticulum (ER).
Functional genomics: we identify essential genes and protein domains from SATAY data using our physics-informed models.

Control theory: we develop models & algorithms that can drive the system to a desired state from given inputs by ensuring a level of control stability and achieving a degree of optimality.
Optimization: we build highly efficient optimization algorithms for the applications in all quantitative disciplines from computer science and engineering to operations research, economics, and biology.
Robotics: we design machines that can help and assist humans using the developed models and algorithms in our group.
Check out our featured projects
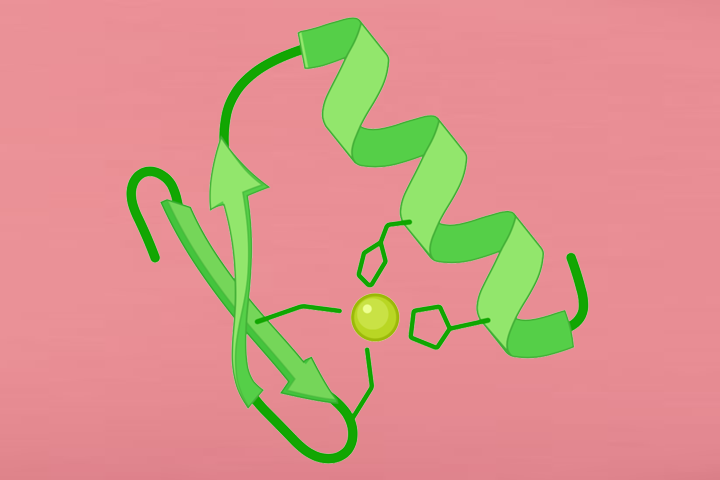
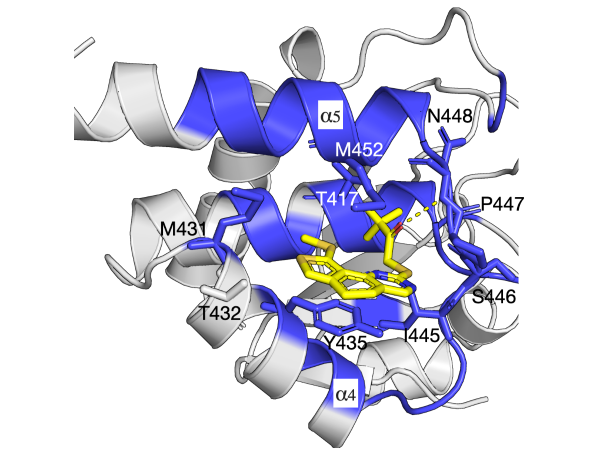
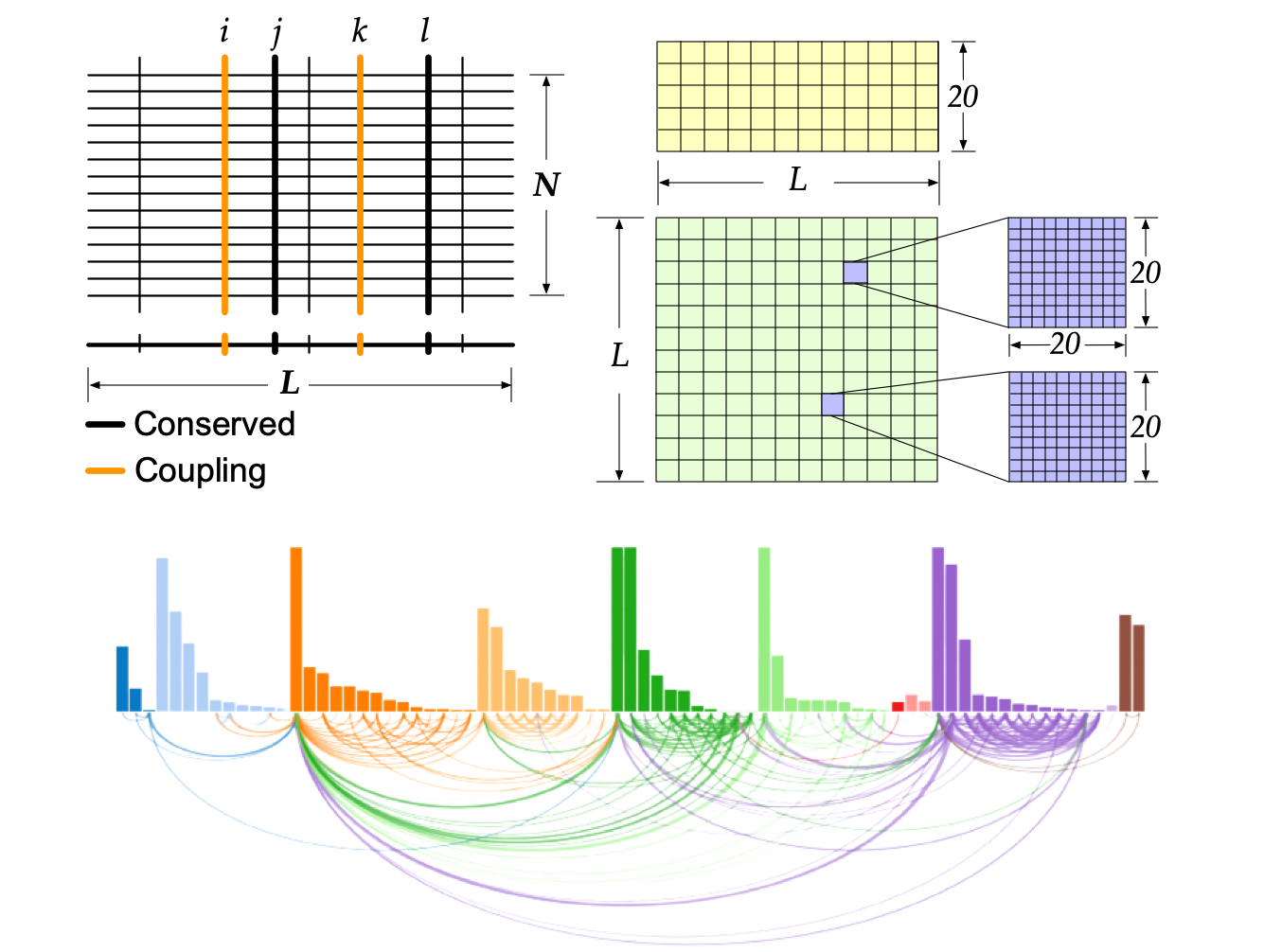
Proteins have highly ordered characterics at primary, secondary, and tertiary levels, and they also play important roles in protein functions.
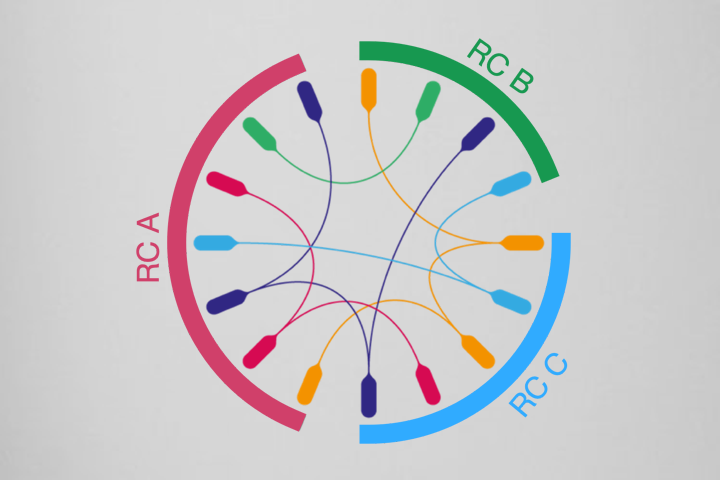
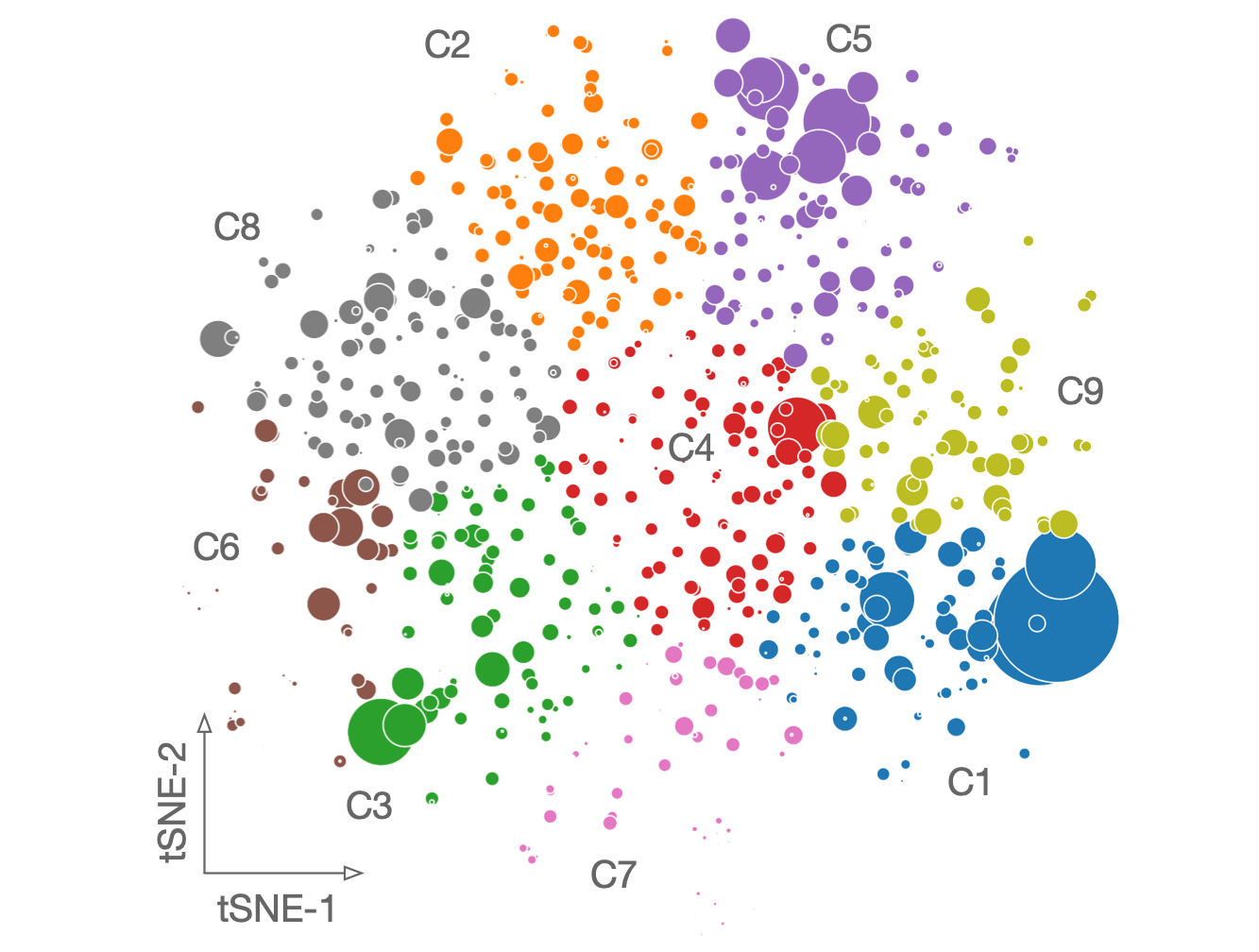
The residue communities, as important evolutionary signatures, would pave a way to engineer protiens for drug development.
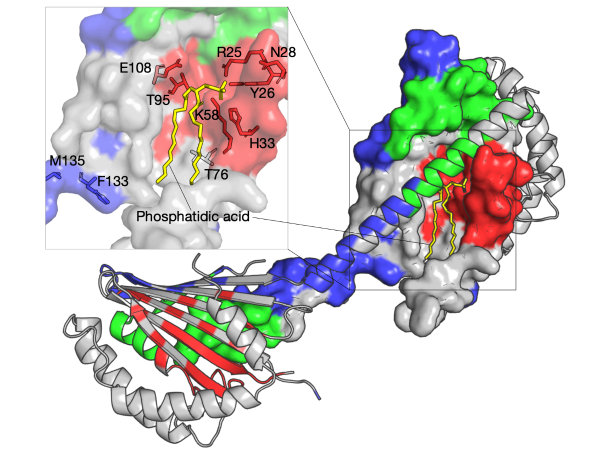
Understanding protein folding, particularly at the atom-level, is often a matter of discovering how it folds, and how it misbehaves in aging or disease.
Awesome works and enjoy





Ema Cerezo
Yong-Chang Xu
University of NÎmes
Get support for the services
AmoAi provides bioinformatics services for and solutions to both academic labs and industries.
Quantify genetic variants for clinical decisions using evolution-informed models.
Design proteins by physics-informed deep learning methods with evolutionary constraints.
Research cutting-edge machine learning methods using information obtained by enforcing physical laws.
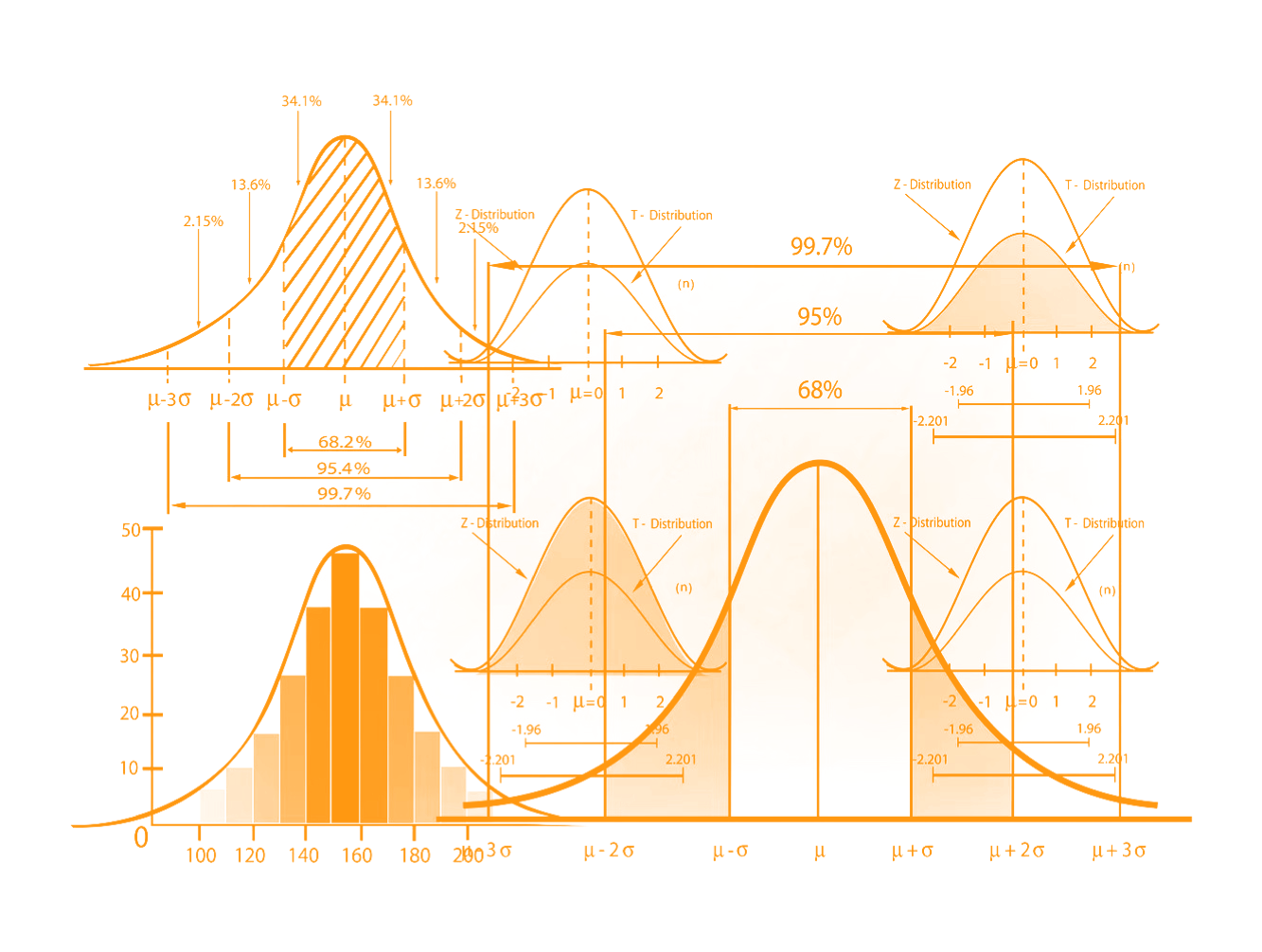
Using understandable & effective graphic representation of data in conveying information
Infer protein function at the residue level from sequences alone using computational approaches.
Feel free to contact us
27 Street, New York, United State.
yaan[DOT]jang[AT]gmail[DOT]com
+075 123 13219
© The Jang Group - All Rights Reserved
July 23, 2020 at 9:32 am